How the Yeasts are Genetically Engineered
The foreign protein or enzyme, which we want to express in our production organism, is first extracted from the original organism, purified and its amino acid sequence is determined. Amino acid sequence of all the enzymes for steviol glycoside biosynthesis pathway is available in database like UniProt.
Next we can back calculate the possible nucleotide sequence in the DNA of the region which codes for the enzyme. There are several bio-informatics tools available for this job. In plant cells finding a gene in this process is not very useful, since the coding region may contain non-sense fragments into it (introns). So it is better to look for the messenger RNA for the particular protein. Once we can figure out the mRNA sequence, and its length, which in turn can give an idea about its molecular weight, we may proceed to the next stage.
From the cells of the tissue where the enzyme is prominently expressed, all the mRNA is collected. The cells are disintegrated first, either mechanically or by ultrasonic treatment. Then total RNA is separated from the homogenate by biochemical methods (Acid guanidinium thiocyanate-phenol-chloroform extraction).
Then mRNA is separated from the total RNA mixture. All the mRNA has a very characteristic “tail” made of adenine residues. The total RNA mix is passed through a matrix with a surface with bound poly thymidine chain. The poly-adenylate tail of mRNA gets bound by the poly-thymidine chain in the matrix and thus, mRNA gets attached to the matrix surface. All the other RNA molecules pass through the matrix freely. Then the mRNA is eluted from the matrix by specific buffer solution and applying little heat.
A strand of DNA, which codes for that particular mRNA is then constructed….just by the reverse process of “transcription” described earlier. The mRNA molecule is incubated with a specifically designed “Primer” (a short nucleotide chain), all the de-oxy nucleotide triphosphates and an enzyme “Reverse Transcriptase”. and a DNA strand, complementary to the mRNA forms. This DNA strand is called single strand “Complementary DNA” or “cDNA”.
The mRNA strands still attached to the cDNAs is then removed by RNA degrading enzymes (RNase). The single strand cDNA molecule is then converted into double strand DNA fragment by incubating it with assortment of nucleotides and another enzyme “DNA Polymerase”.
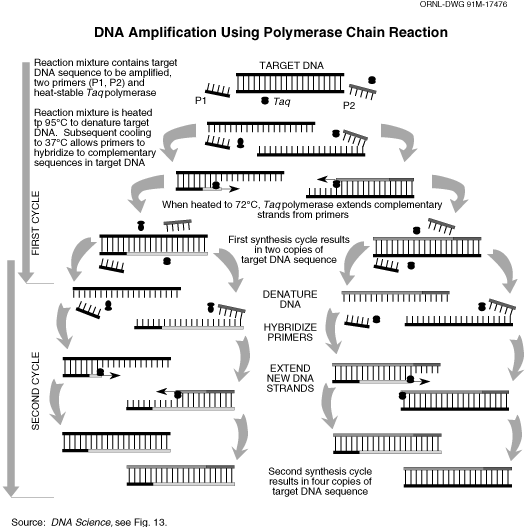
This operation results into transcription of all the mRNA coding different proteins. Now to separate our genes of interest, that specific gene is “amplified” by a process called Polymerase Chain Reaction (PCR). For PCR, we need short nucleotide chains which bind at one end of the cDNA length corresponding to the gene. For that, we need to know the nucleotide sequence of the both ends of the cDNA, so that we can design a short stretch of DNA with complimentary bases which will specifically bind at the forward and reverse end of the gene. For Steviol glycoside biosynthesis pathway, we already know the full nucleotide sequence of the all the genes, designing of those “primers” are not difficult. PCR makes several copies of the DNA sequence flanked by these primers.
The amplified DNA thus made is separated by gel electrophoresis. Then, if required, specific promoter sequence and start/stop codons is attached to the DNA fragment.
The specific gene thus made, is then to be transferred into the yeast cell. For this gene transfer, the DNA fragment for the Gene is to be packed in a “vector”. The vectors are often short pieces of circular DNA with capability of replicating freely in a microbial cell. These circular DNA pieces are called “Plasmids” Along with it’s the codes for its own replication, the vector may contain the desired gene to be introduced, some markers and “Reporter genes” to prove that the vector actually entered into the target organism and functioning. There are two major types of plasmid vectors for yeast. One is Yeast integrative plasmid (Yip). Genes in these plasmids gets integrated into the yeast genome and imparts very stable inheritable characters. The other type is Yeast Replicative Plasmid (YRp), which does not get integrated into the chromosomal DNA but replicates independently in the yeast cell. These types of plasmids are prone to loss during yeast reproduction and the genetic characteristics imparted by these vectors are less stable.
The yeast cells are incubated with those vectors along with some inorganic salts, which facilitate the entry of the vectors in the yeast cells. The uptake of vectors by the yeast cells and formation of yeast cells expressing the transferred genes is called “Transformation”. The cells which shows the expression of the genes are called transformed cells, and they are selected on the basis of easily identifiable genetic markers or reporter genes transferred to them by the vector along with the gene coding for the target enzyme.
The transformed cells are then isolated, there pure culture is made and used as production strain.